
There are inherent efficiency losses with two-stage approaches, even when adequate models are used in each stage ( Gogel et al., 2018). This can be contrasted with two-stage approaches in which variety means from the separate analysis of each environment (stage 1) are used as “data” in a subsequent MET analysis (stage 2). All of this is achieved using a one-stage approach in which a single statistical analysis is conducted using the individual plot data combined across environments. Furthermore, the FA component of the model consistently provides a good fit to the data and allows quantification and interpretation of VEI.
LEAST SQUARE MEANS ASREML TRIAL
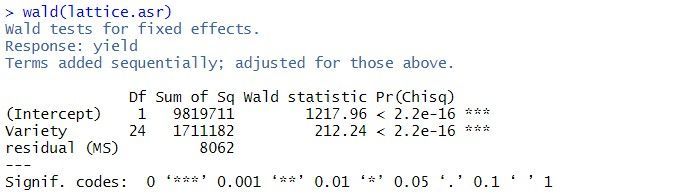
Key benefits are the ease with which incomplete data (not all varieties grown in all environments) can be handled, the ability to appropriately account for individual trial designs and the ability to include information on genetic relatedness, either through ancestral (pedigree) or genomic (marker) data. It is widely known that there are numerous advantages in analyzing MET datasets using a linear mixed model (LMM) approach in which a factor analytic (FA) variance structure is assumed for the variety effects in individual environments (see Smith et al., 2005, 2021 Gogel et al., 2018, for example). They are an important component of identifying superior varieties as they allow an assessment of variety by environment interaction (VEI), that is, the differential performance of varieties in response to a change in environment.
LEAST SQUARE MEANS ASREML SERIES
Plant breeding multi-environment trials (METs) comprise series of variety trials conducted at a range of geographic locations and typically across several years (synonymous with seasons). The application to FALMMs which include information on genetic relatedness is the subject of a subsequent paper. The ideas are introduced in this paper within the framework of FALMMs in which the genetic effects for different varieties are assumed independent. The latter is aided with the use of a new graphical tool called an iClass Interaction Plot. These predictions can then be used not only to select the best varieties within each iClass but also to match varieties in terms of their patterns of VEI across iClasses. Given that the environments within an iClass exhibit minimal crossover VEI, it is then valid to obtain predictions of overall variety performance (across environments) for each iClass. These groups are consequently called interaction classes (iClasses).


This is addressed in the current paper by fitting a factor analytic linear mixed model (FALMM) then using the fundamental factor analytic parameters to define groups of environments in the dataset within which there is minimal crossover VEI, but between which there may be substantial crossover VEI.



 0 kommentar(er)
0 kommentar(er)
